Help/About
The Metabolism and Transport Database (Metrabase) is a cheminformatics and bioinformatics resource that provides structured data about the interactions between proteins and chemical compounds related to their metabolic fate and transport across biological membranes.
The aim is to build a comprehensive resource providing high quality structural, physicochemical and biological data that entails minimal processing load for users. The data held in this database can be utilised to infer the relationships between transporters/enzymes and their ligands. The database also contains compounds that were experimentally found not to be substrates (as well as non-inhibitors and non-inducers), which makes it a valuable resource for building predictive models based on the characteristics of both the positive and the negative class.
The initial focus of the project was on substrates and non-substrates of transport proteins, and as such the related activity records have undergone an additional level of annotation and checking in comparison to the inhibition- and induction-related data.
Metrabase v1.1
20 transporters and 13 CYPs: 3437 compounds, 11662 interaction records, 1209 literature references
20 transporters: 3306 compounds, 11154 interaction records, 1175 literature references
13 CYPs: 213 compounds, 508 interaction records, 37 literature references
-
Support and feedback
Please contact us at metrabase@ch.cam.ac.uk. -
How to cite
Please cite: Mak L, Marcus D, Howlett A, Yarova G, Duchateau G, Klaffke W, Bender A, Glen RC: Metrabase: a cheminformatics and bioinformatics database for small molecule transporter data analysis and (Q)SAR modeling. Journal of Cheminformatics 2015, 7:31.
http://www.jcheminf.com/content/7/1/31. -
Licensing
Metrabase is licensed under a Creative Commons Attribution-ShareAlike 4.0 International License. However, the integrated data retains the licensing of the original data sources. The TP-Search and ChEMBL activity records may have been modified and augmented, while the expression records taken from The Human Protein Atlas were included unmodified. The 'datasource_id' and 'datasource_version' fields indicate the source of each relevant Metrabase record. -
Data sources
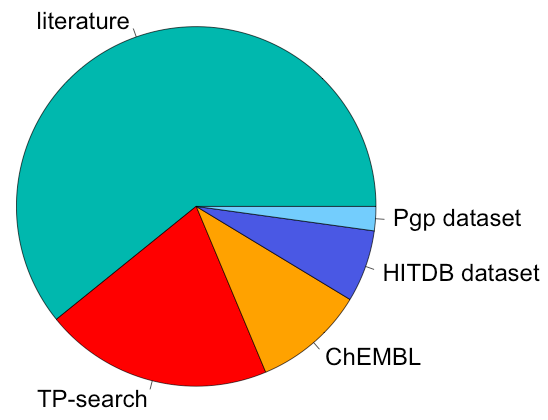
Data was manually extracted from published literature and combined with other available resources: TP-Search, ChEMBL, Human Protein Atlas, DrugBank and UniProt. Data from two recently published papers (PMID: 23269503 (HITDB) and the Pgp dataset from PMID: 22595422) were also integrated. The pie chart below depicts proportions of interaction records per data source. Please note that all the data was extracted from the published literature: where datasource='literature', it only means that the data was extracted by us. SciFinder and ChemSpider were used to check the correctness of chemical structures and names. Their compound IDs are provided as external IDs: CAS Registry Numbers (CASRNs) are provided only if ChemSpider IDs (CSIDs) were not available. CAS Registry Number is a Registered Trademark of the American Chemical Society.
-
Search by protein
Users can select a protein from the drop down menu and one or more action types, such as substrate and non-substrate. This will list the compounds that were found to be substrates or non-substrates of the selected protein. The output also includes cell lines and experimental values for different quantities, such as uptake, efflux, Michaelis-Menten constants (Km and Vmax), Ki and IC50, where available. -
Action types
The following action types are defined: substrate, non-substrate, inhibitor, non-inhibitor, stimulator, inducer, non-inducer, repressor and binder. Substrates and non-substrates are compounds that were experimentally found to be or not to be transported/converted by transporters/enzymes, respectively. Compounds were categorised as substrates or non-substrates according to the results presented in the publication that the data was extracted from, and no further evaluation was carried out on our side. The 'action_strength' field of the 'activities' table indicates weak substrates, but please note this field was not done comprehensively. Action types related to protein expression include inducer (increased level of expression), non-inducer (unchanged expression levels) and repressor (decreased level of expression). Action types related to protein activity include inhibitor (decreased activity), non-inhibitor (unchanged activity) and stimulator (increased activity). Care must be taken with respect to the current status of the inhibition records, since depending on the measurement threshold (e.g. percentage inhibition) some of the compounds annotated as inhibitors can be regarded as non-inhibitors or vice versa. Finally, where the action type did not fall into any of these categories, but the molecule was found to bind to the protein, its action type was set to binder.
Action types Protein activity
(transport or catalysis)substrate
non-substrateCompound activity
(affecting protein activity/expression)inhibitor/repressor (negative modulators)
stimulator/inducer (positive modulators)
non-inhibitor/non-inducer (inactive compounds) -
Expression data
Metrabase also contains information about protein expression levels across healthy human tissues. Part of this data is based on immunohistochemistry using tissue microarrays and comes from the normal_tissue.csv file of the Human Protein Atlas v9.0. All the other records contain data that we extracted from the literature. The levels of expression for non-HPA records (i.e. where ‘ref_id’ is not null): expressed (if the level has not been specified), none, none-low, low, low-medium, medium, medium-high and high. -
Protein list
The numbers of activity records and compounds are provided per protein. Links are given to UniProt and HGNC web pages. Sequences for these proteins can be downloaded in the FASTA format. -
Download
The database and other related files can be downloaded from here. -
 Acknowledgement
Acknowledgement
We thank Unilever for funding and supporting the project.
We thank Eisai Ltd. for their contribution toward the summer student funding.
We are also grateful to all authors of freely available scientific resources and software tools without which this work would have been much harder. These also include, not so far mentioned, MySQL Community Server, Open Babel, JabRef and the NCBI databases and services.
 We thank ChemAxon for an academic licence.
We thank ChemAxon for an academic licence.
-
Credits
Metrabase was developed by Lora Mak in collaboration with David Marcus, Andreas Bender and Robert C. Glen at the Unilever Centre for Molecular Sciences Informatics and Galina Yarova, Guus Duchateau and Werner Klaffke at Unilever, with the much appreciated help from the following (at the time) 2nd and 3rd year undergraduate students of the University of Cambridge: Claire Dickson, Joseph Dixon, Ivan Lam, Richard Lewis, Callum Picken, Claudia Pop, Heyao Shi, Emma Stirk, Yasmin Surani, Paddy Szeto, Nathaniel Wand, Julian Willis and Jing Xiangyi.
The first version of Metrabase's web interface (a Java application) was developed by Andrew Howlett at the Unilever Centre for Molecular Science Informatics.
Metrabase was realised and is being maintained in the Glen group.


